SIR model
\( \frac{\mathrm{d}}{\mathrm{d}t}I=r\beta I\frac{S}{N}-\gamma I \\ \frac{\mathrm{d}}{\mathrm{d}t}S=-r\beta I\frac{S}{N} \\ \frac{\mathrm{d}}{\mathrm{d}t}R=\gamma I\)
View all models and descriptions: Infectious disease model simulation and theoretical trends
clear
figure
N = 300; % total population
I = 3; % infector state == 1
S = N-I; % susceptible state == 0
R = 0; % recovered state == 2
r = 2; % number of infected persons contacting susceptible persons
B = 0.05; % probability of infection
y = 0.03; % recovery probability
days = 500;
Dynamic = false; %Set to true to show the dynamic process
Sdata=[];
state=zeros(1,N);
index=randperm(N,I);
state(index)=1;
if Dynamic
axis([0 N 0 N]);
hold on
end
for i=1:days
xpos=randperm(N);
ypos=randperm(N);
for j=1:N
if state(j)~=1
continue
end
if rand()<=y
state(j)=2;
continue
end
dis=sqrt((xpos-xpos(j)).^2+(ypos-ypos(j)).^2);
l=0;
for k=1:N
if state(k)==1 || state(k)==2
continue
end
l=l+1;
peo(l).dis=dis(k);
peo(l).num=k;
end
T = struct2table(peo);
sortedT = sortrows(T,'dis');
sortedS = table2struct(sortedT);
tp=sortedT{1:r,2};
for k=1:length(tp)
if rand()<=B
state(tp(k))=1;
end
end
end
Idata(i)=length(find(state==1));
Sdata(i)=length(find(state==0));
Rdata(i)=length(find(state==2));
%a=-8;
%b=8;
%xpos=xpos+ a + (b-a).*rand(1,N);
%ypos=ypos+ a + (b-a).*rand(1,N);
if Dynamic && rem(i,10)==0
phd=scatter(xpos,ypos,[],state,"filled");
title(['Day ' num2str(i)])
drawnow
delete(phd)
end
end
figure
set(gcf,'visible',true)
days=length(Sdata);
plot(1:days,Sdata,1:days,Idata,1:days,Rdata)
hold on
T = 1:days+100;
for idx = 1:length(T)-1
if S(idx)<0
S(idx)=0;
elseif S(idx)>N
S(idx)=N;
end
if I(idx)<0
I(idx)=0;
elseif I(idx)>N
I(idx)=N;
end
if R(idx)<0
R(idx)=0;
elseif R(idx)>N
R(idx)=N;
end
S(idx+1) = S(idx) - r*B*S(idx)*I(idx)/N;
I(idx+1) = I(idx) + r*B*S(idx)*I(idx)/N - y*I(idx);
R(idx+1) = R(idx) + y*I(idx);
end
plot(T,S,T,I,T,R);
legend ('simulated susceptible person', 'simulated infectious person', 'simulated recovered person', 'susceptible person', 'infected person', 'recovered person')
xlabel ('day'); ylabel ('number of people')
title ('SIR model')Code language: Matlab (matlab)The final screenshot of the simulation is as follows (the dynamic process is not given):
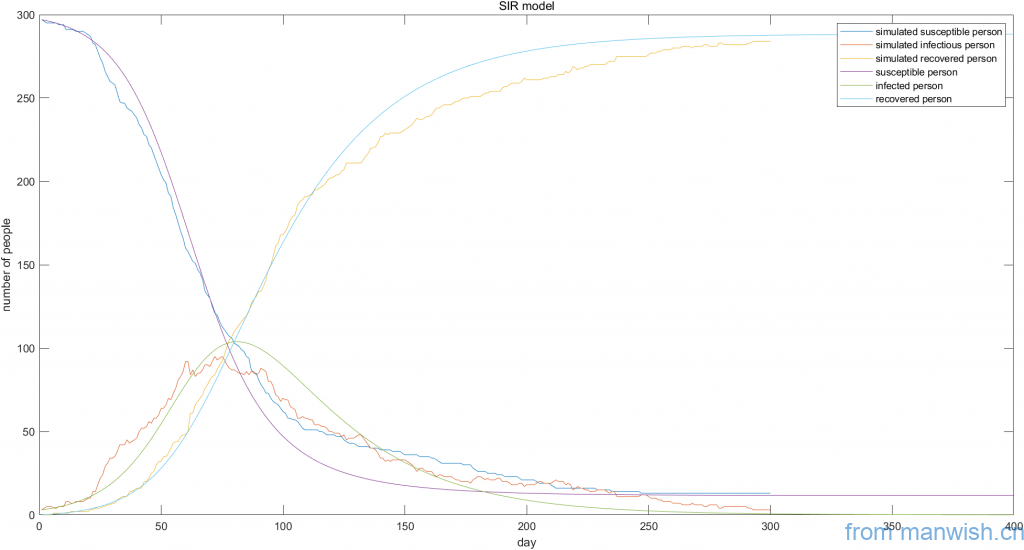
Unless otherwise stated, the articles are original.
Title of this article:Dynamic simulation and theoretical curve of SIR virus model
Hyperlink to this article:https://manwish.cn/en/article/dynamic-simulation-and-theoretical-curve-of-sir-virus-model-en.html
Title of this article:Dynamic simulation and theoretical curve of SIR virus model
Hyperlink to this article:https://manwish.cn/en/article/dynamic-simulation-and-theoretical-curve-of-sir-virus-model-en.html